Technology
The NGS method analyses all chromosomes of the embryo and detects mosaicism and unbalanced chromosomal rearrangements
Embryo screening in ART to increase the chances of successful implantation, pregnancy and birth.
PGT-A or preimplantation genetic testing for aneuploidy is the screening of embryos in an assisted reproductive technology cycle for chromosomal abnormalities before they are transferred into the uterus to increase the likelihood of achieving a successful pregnancy.
PGT-A was formerly known as PGS, preimplantation genetic screening
20%of human eggs
9%of human spermatozoa
In natural gametogenesis, approximately 20% of human eggs and 9% of human spermatozoa are thought to be aneuploid*. Aneuploid gametes involved in the fertilisation process lead to the development of aneuploid embryos, often resulting in implantation failure or pregnancy loss.
*Martin RH. Meiotic errors in human oogenesis and spermatogenesis. Reprod BioMed Online 2008;16(4):523-31
38%–76%of spontaneous abortions are associated with chromosomal abnormalities
According to world and Russian statistics, the prevalence of chromosomal abnormalities detected in spontaneous abortion ranges from 38% to 76%*. The frequency of aneuploidy in embryos in ART cycles increases with maternal age, reaching 80% in patients over 40 years of age**.
* Chiryaeva O.G. et al., 2012, Volkov A.N. et al. 2017, Carp H, et al., 2001, Rosa Russo et al., 2016)
**Macklon NS et al. Conception to ongoing pregnancy: the “black box” of early pregnancy loss. Hum Reprod Update 2002;8:333-343
38%–73%of embryos in ART cycles contain aneuploid cells
Recent large studies of human embryos have shown that 38-73% of embryos in ART cycles contain aneuploid cells. The differences in the statistics are mainly due to the different average age of the oocytes
Le Thi Bich Phuong et all. Selecting euploid embryo for transfer by preimplantation genetic testing for aneuploidy improved clinical outcomes in patients with advanced maternal age DOI : 10.15419/bmrat.v6i12.581
X Viñals Gonzalez et all, Euploid Blastocysts Implant Irrespective of Their Morphology After NGS-(PGT-A) Testing in Advanced Maternal Age Patients, 2019.
PGT-A is becoming one of the most valuable tools for increasing the success of assisted reproductive technology pregnancies. As indicators of mosaicism can be quantified by NGS, mosaic embryos on non-viable chromosomes can be considered for transfer when euploid embryos are not available.
Increasing the frequency of implantation
Up to an average of 50-69%, data varies according to age group and associated clinical findings
Simon AL, et al, Pregnancy outcomes from more than 1,800 in vitro fertilization cycles with the use of 24-chromosome single-nucleotide polymorphism-based preimplantation genetic testing for aneuploidy. Fertil Steril. 2018
Reducing the rate of non-developing pregnancies
In the general population, 25% of all clinical pregnancies end in miscarriage. About 50% of sporadic early miscarriages are due to chromosomal defects. The risk of miscarriage decreases when an euploid embryo is transferred.
Early miscarriage: diagnosis and management, clinical guidelines.
Reducing the time to gestation
The time to gestation with PGT-A is reduced from 15 weeks to 8 weeks.
Rubio et al., In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: a randomized, controlled study. Fertil Steril. 2017
Reducing the risk of complications in multiple pregnancies
The high rate of multiple pregnancies is a consequence of multiple embryo transfer in ART programmes. Multiple pregnancies can also occur as a consequence of a naturally occurring pregnancy. According to the 2015 RAHR ART Registry, the rate of multiple pregnancies in the IVF and ICSI programmes was 19.7% of all births, 15.2% following an unfrozen embryo transfer, 20.4% in the Oocyte Donation programme and 25.2% in the surrogacy programme (91).
Selective transfer of 1 embryo into the uterine cavity is recommended to prevent multiple pregnancies.
ART and artificial insemination, clinical guidelines
After the vitrification period, euploid embryos are transferred, allowing time for screening for chromosomal abnormalities and preparing the endometrium for implantation.
In all of these patient groups, an increased incidence of chromosomal abnormalities in embryos is suspected.
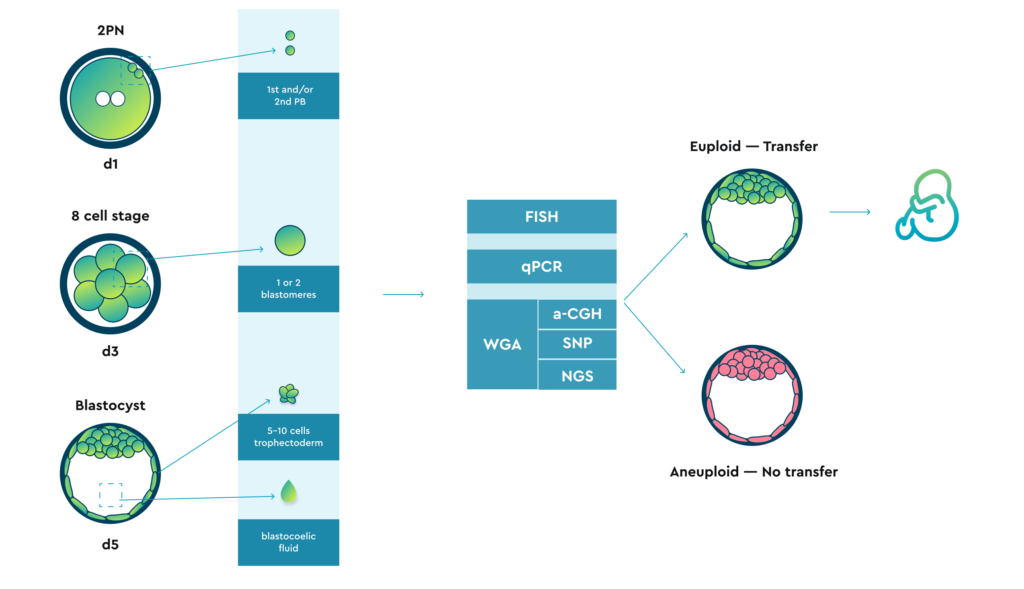
Mosaicism can exist in cells that are part of the trophectoderm. The ability to identify mosaicism within the trophectoderm posed a significant challenge and the following approaches have been implemented to overcome this problem:
Current diagnostic technology tests cells intended to become placenta, not cells of the intracellular mass that differentiates into the foetus. Biopsy of the intracellular mass is not recommended because of concerns about further fetal differentiation.
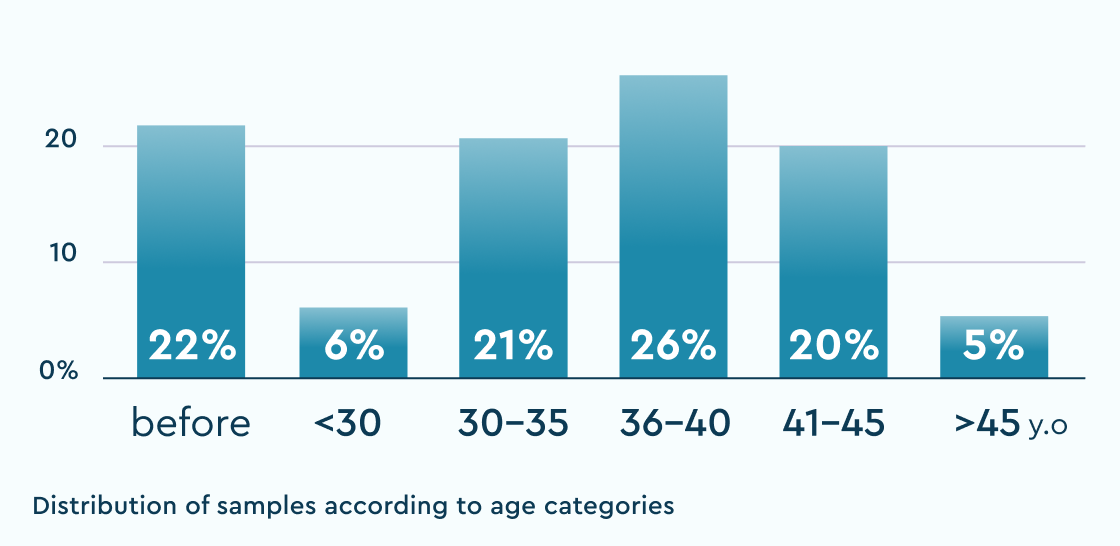
~8000samples
% of the norm
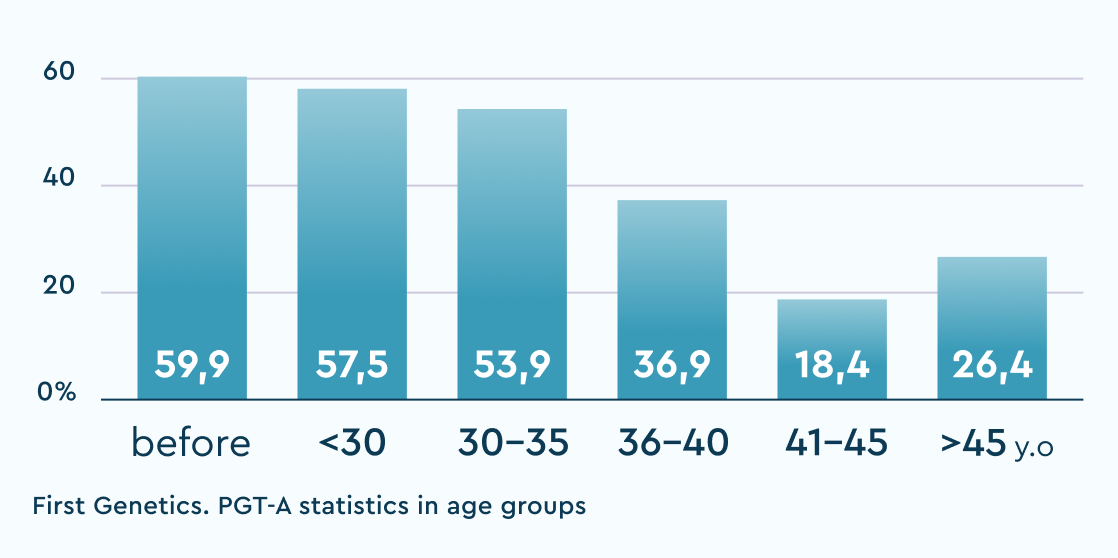
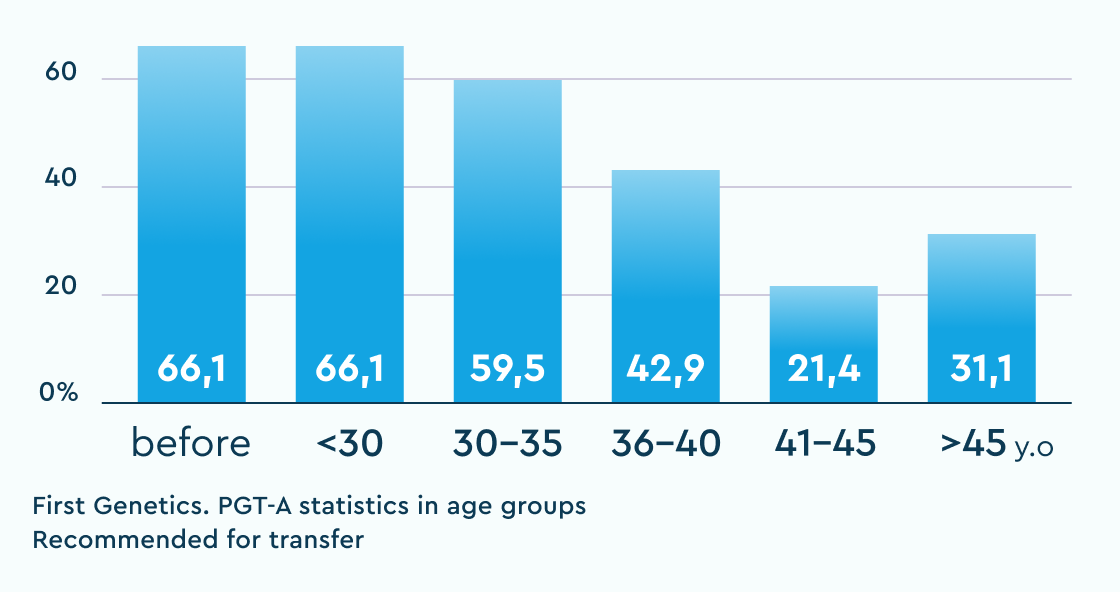
Recommended for transfer (N+mos)
Isolated segmental anomalies (%)
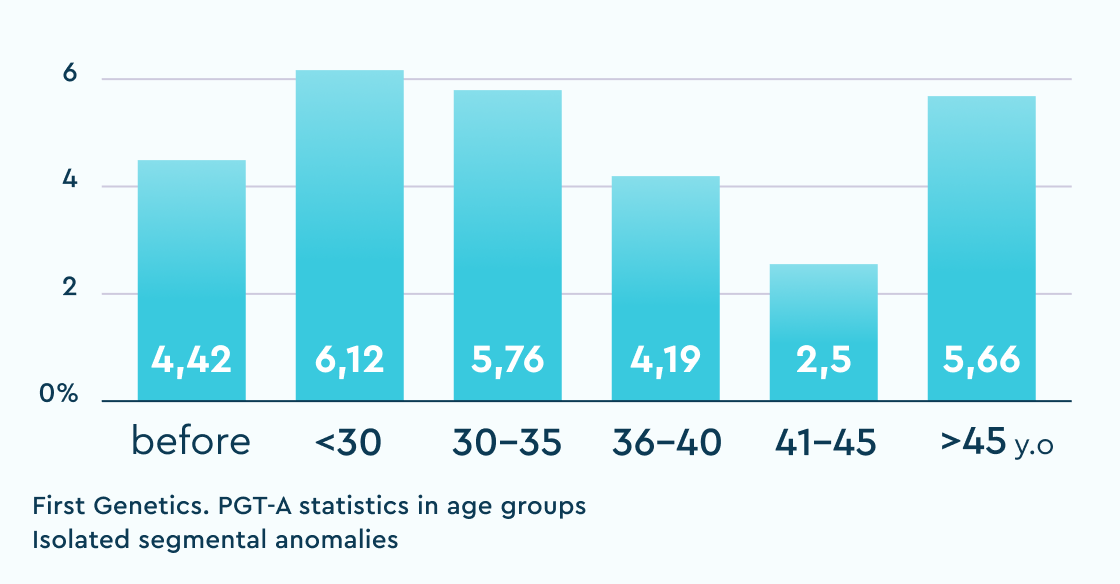
The table shows the results of "overachievers" clinics in Russia with whom we have an established cooperation and no doubts about the "correctness" of their "embryological" protocol. "Underachievers" are single clinics where deviations from the usual statistics were observed and solutions were proposed to audit and search for potential causes of the violations.
Based on data from the transfer of more than 700 euploid embryos, it has been shown that negative predictors of outcome can be:
the number of pregnancy failures
embryo biopsy on day 6 (compared to day 5)
elevated BMI
Fazilet Kubra Boynukalin et al.,Parameters impacting the live birth rate per transfer after frozen single euploid blastocyst transfer, 2020
for sensitive detection of mosaicism and segmental abnormalities of all chromosomes
1.5 calendar days
(upon agreement)
7 calendar days
12 working days
at every stage of testing
from anywhere in Russia
A biopsy can potentially be performed at three different stages:
Additionally, there are reports in the literature of attempts to aspirate a small volume of blastocoietic fluid.
All types of the previously mentioned biomaterial can potentially be analysed using different technologies. However:
Chromosome analysis has been used in clinical practice since the mid-1990s.
FISH FISH fluorescent hybridisation
1990s
Initially, this diagnostic strategy was performed using fluorescent in situ hybridisation (FISH), which consisted of plating a single embryo cell at the cleavage stage (blastomer) on a slide and hybridising its DNA with chromosome-specific fluorescent probes. The limited number of chromosomal probes examined also meant that some aneuploidies remained untested, leading to the transfer of undetected aneuploid embryos.
In the mid-2000s, it became clear that these shortcomings and diagnostic unreliability, combined with the negative consequences of biopsy at the cleavage stage, jeopardised the clinical outcomes of patients for whom PGT-A had been performed, prompting the need for a safer, more reliable and more accurate strategy.
CCS methods CCS comprehensive chromosome screening
>98% the accuracy that all CCS platforms have shown
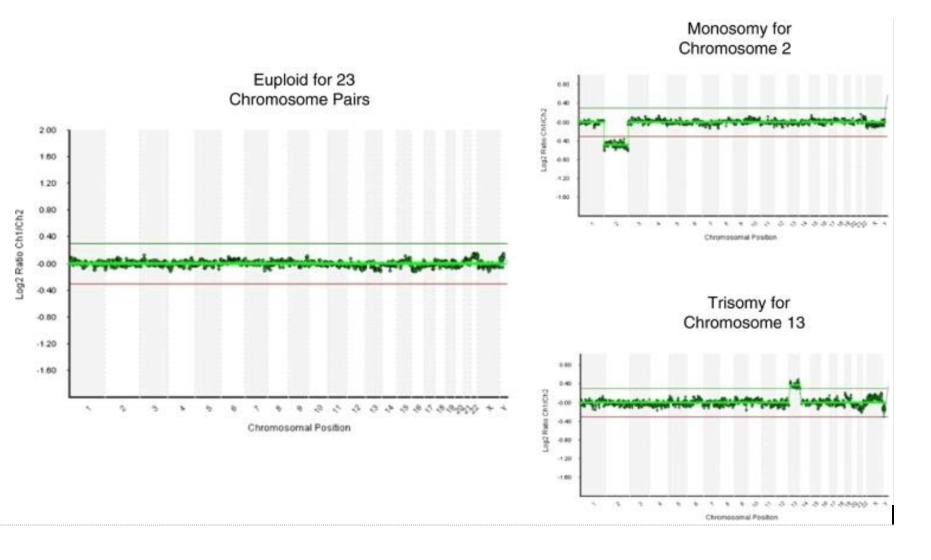
Subsequently, the development of comprehensive chromosomal screening (CCS) technologies, including comparative genomic hybridisation (aCGH), single nucleotide polymorphism arrays (SNP arrays) and quantitative polymerase chain reaction (qPCR), have provided significant improvements in the clinical application of PGT-A.
When tested on a single cell from fibroblast cell lines with a known karyotype, all CCS platforms showed an accuracy above 98% for the detection of complete aneuploidies.
All platforms that can analyse all 23 pairs of chromosomes have comparable effectiveness in detecting aneuploidy of an entire chromosome, but differ from each other
aCGH, SNP and massively parallel sequencing (MPS, NGS) require a first step of whole genome amplification (WGA).
FISH and qPCR do not require a pre-amplification step.
NGS provides greater accuracy in assessing subchromosomal anomalies (e.g. segmental aneuploidies) than previous CCS methods.
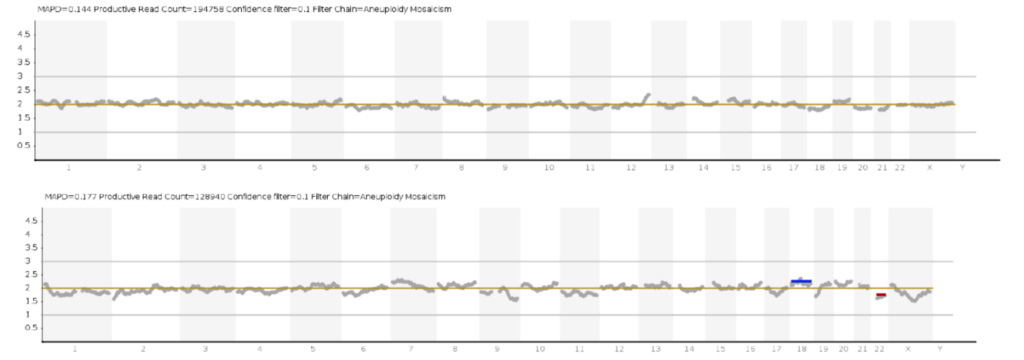
In addition, NGS is used to detect chromosomal mosaicism (when two karyotypically different cell populations coexist in the same embryo).
However, it is important that the detection of low and high levels of mosaicism (e.g. 20 and 80%, respectively) should be interpreted with extreme caution, as it is quite difficult to distinguish biological findings from the technical features of sequencing that particular sample at these values of mosaicism. This point is extremely important, especially for patients with a limited number of embryos.
In addition, some diagnostic platforms can also test for chromosomal aberrations and single gene mutations simultaneously.
Quantitative PCR (qPCR) or real-time PCR (PCR-RV) is a polymerase chain reaction that can identify aneuploidy of an entire chromosome by determining the copy number of each chromosome analysed.
This is achieved by comparing three or four locus-specific amplicons along each chromosome with a reference gene localised on the same chromosome.
This analysis can identify aneuploidy for all 23 chromosome pairs in a short time (4-12 hours; depending on the number of samples analysed), and due to the labour-intensive nature of the process in the absence of automation, there is a high risk of error due to human error.
qPCR cannot identify structural chromosomal aberrations but can identify triploidy.
As qPCR does not detect genotype, it cannot identify uniparental dysosomes. A separate experiment plan can be devised to determine the number of mitochondrial copies.
There are two main types of microarray available for genetic testing. These are single nucleotide polymorphism arrays and comparative genomic hybridisation.
For both microarray platforms, trophectoderm cells must be lysed and amplified using some type of DNA amplification protocol that ensures that the entire genome is covered. As with any genetic test, the quality of the diagnostic result begins with the quality of the amplified DNA sample.
SNPs (single nucleotide polymorphisms) are pairs of single nucleotides (A, T, C or G) in genomic DNA that vary widely within a given species. In the context of PGT-A, the SNPs assessed are usually in non-coding parts of the genome.
After WGA, the embryonic DNA is fragmented and hybridised with an SNP microarray, which contains probes for more than 300,000 different SNP sites across the genome.
After hybridisation, a chain lengthening and staining step is performed. A/T nucleotides at the SNP site are labelled with red fluorochrome, and G/C nucleotides at the SNP site are labelled with green fluorochrome.
By measuring the fluorescence intensity from red to green at each SNP site in the array, it is possible to simultaneously determine the genotype of more than 300,000 SNPs in each sample and compare the results with the reference human genome map.
The data arrays can identify whole chromosome aneuploidy and can also identify approximately 250 common structural chromosomal aberrations across the genome.
However, there are hundreds of other chromosome structural abnormalities below the resolution of the 300 000 SNP arrays used for PGT-A, which may play an important role in implantation, miscarriage or the birth of a child with a serious genetic syndrome. As genotype information is provided, these SNP arrays have limited ability to identify triploidy, but can identify uniparental dysomy.
SNP microarrays can also identify mosaicism if a sufficient number of trophectoderm cells have been analysed. One limitation of SNP microarrays used for PGT-A is the inability of their algorithm to identify the number of copies when the husband and wife are blood relatives.
CGH (aCGH) microarrays are less dense than SNP microarrays. The aCGH chips used for PGT-A have approximately 4000 markers (which are run in a double) located throughout the genome.
aCGH is a protocol for identifying the copy number ratios of a clinical sample to a female or male reference genome. The assay can be performed in a shorter time compared to SNPs (in 12-15 hours).
This is a significant advantage over the SNP microarray analysis, which takes around 30-40 hours to complete. Genotypes that are identified in SNPs are not detected in this type of analysis, which means that aCGH cannot distinguish karyotype 46, XX from 69,XXX or 46, XY from 69,XXY. In addition, aCGH cannot detect uniparental dysomy. aCGH, which is used for PGT-A in all commercial laboratories, can only identify whole chromosome aneuploidy and is not designed or validated to identify structural chromosomal aberrations. Even if aCGH microarray is validated for mosaic samples, it should be understood that the ability to detect mosaicism in trophectoderm samples is still limited.
According to some reports, the error rate for aCGH is approximately 15-30%*.
*Capalbo A, Treff NR, Cimadomo D, et al. Comparison of array comparative genomic hybridization and quantitative real-time PCR-based aneuploidy screening of blastocyst biopsies. Eur J Hum Genet: EJHG. 2015;23:901–6. doi: 10.1038/ejhg.2014.222
In other validation studies, when comparing NGS and CGH, the concordance rate is as high as 99%**.
** data by Paul R. Brezina, on 400 samples
Next Generation Sequencing (NGS) is a technology that requires very "careful" DNA amplification to reduce the likelihood of artefacts during the amplification process.
However, after DNA amplification, in most cases artefacts can be identified and removed by bioinformatics analysis.
There are currently two main platforms used for PGT-A. These are Illumina's MiSeq and Thermo-Fisher Scientific's S5. S5 is an improved version of its predecessor PGM.
After DNA amplification, approximately 50 ng of each DNA sample is enzymatically cleaved into millions of fragments and combined to prepare a library. Library preparation is the process by which all the DNA fragments are 'cross-linked' with an adapter and a unique identifier, the barcode.
A well-prepared library creates a representative objective representation of nucleic acids and is crucial for accurate molecular analysis.
After library preparation, either a bridging PCR step is performed. For MiSeq, sequencing takes place by synthesis based on fluorescence detection, which is recorded using an optical camera.
The platform allows up to 24 samples to be analysed simultaneously.
Once the library has been prepared, either the emulsion PCR step is performed. S5 uses an ion-sensitive field effect in which a signal is detected as the hydrogen ion is released, which occurs whenever a nucleotide triphosphate is incorporated by DNA polymerase during sequencing.
The release of a proton causes a slight shift in pH, which is recorded by a sensitive sensor.
The platform is scalable and can analyse 16, 24 or 96 samples simultaneously.
Both MiSeq and S5 sequence the whole genome and compare the sequencing data with the reference human genome.
Both platforms allow to perform the analysis (from DNA amplification to final report) in 12-16 hours.
After sequence analysis, significant differences arise between MiSeq data analysis and S5 data analysis.
MiSeq DNA passes the first round of quality assurance indicators, followed by analysis using BlueFuse software.
MiSeqfrom Illumina
Analysis on S5 passes the first round of quality assurance indicators using the Torrent Browser, followed by detailed analysis using the Ion Reporter software.
S5 и PGM from Thermo-Fisher Scientific
There are differences in the data analysis of the two platforms. The main point to note is that the bioinformatic data analysis algorithm in Ion Reporter is flexible, customisable to different tasks and available for detailed study. Detailed information can be found in the brochure:
1. Poor sample quality and the emergence of amplification artefacts. Poor sample quality (e.g. degraded DNA) and problems in the DNA amplification step result in increased overall noise and peaks.
2. The quality of whole genome amplification can be affected by a number of factors:
These factors tend to be more prominent in single-cell biopsy samples than in trophoectoderm biopsy samples, where more source material is available.
3. insufficient reads can also lead to a noisy profile and the assignment of a false number of copies.
4. How time-consuming certain steps in some manufacturers' protocol are (e.g. washing each sample on magnetic particles can lead to over-drying and deterioration of the sample).
It is very important when analysing multiple samples that the DNA concentrations of the samples in the pooled library are balanced.
Amplification causes 'skewing' towards certain fragments — thus causing the problem of uneven coverage. The solution is:
Duplicated reads can over-represent the amplification artefact. The solution is:
When processing the PGT-A results, laboratory technicians take into account the need to detect small segmental abnormalities.
If mosaicism (for complete chromosomes or segmental abnormalities) is detected, specialists make a clear recommendation as to whether a particular embryo can be considered for transfer with the patient's informed consent.
In this scenario, patients should consult their doctor about the consequences of such a transfer.
Separate and special attention is required when considering the possibility of non-lethal aneuploidy transfer (along sex chromosomes 47,XXX, 47XXY, 47XYY).
Собственное IT решение «Личный кабинет» — еще одно преимущество сотрудничества с First Genetics.
Для врачей клиник-партнеров предоставляется доступ в личный кабинет, который позволяет отслеживать каждый этап исследования.
Решение разработано при помощи:
Ознакомится с функционалом решения можно в видео инструкции.
The NGS method analyses all chromosomes of the embryo and detects mosaicism and unbalanced chromosomal rearrangements
Years of experience in genetics, laboratory diagnostics and bioinformatics
All data is strictly confidential and cannot be passed on to third parties
You can get an online consultation regarding the results of test
Extensive control at each stage of testing
Free delivery of biomaterial across Russia